Exploring NWB Files

NWB Widgets
A library of widgets for visualizing NWB data in a Jupyter notebook (or lab). The widgets make it easy to navigate through the hierarchical structure of the NWB file and visualize specific data elements.
Neurosift
Interactive neuroscience visualizations in the browser. Neurosift caters to both individual users through its local mode, allowing the visualization of views directly from your device, as well as a remote access function for presenting your findings to other users on different machines.
NWB Explorer
A web application developed by MetaCell for reading, visualizing and exploring the content of NWB 2 files. The portal comes with built-in visualization for many data types, and provides an interface to a jupyter notebook for custom analyses and open exploration.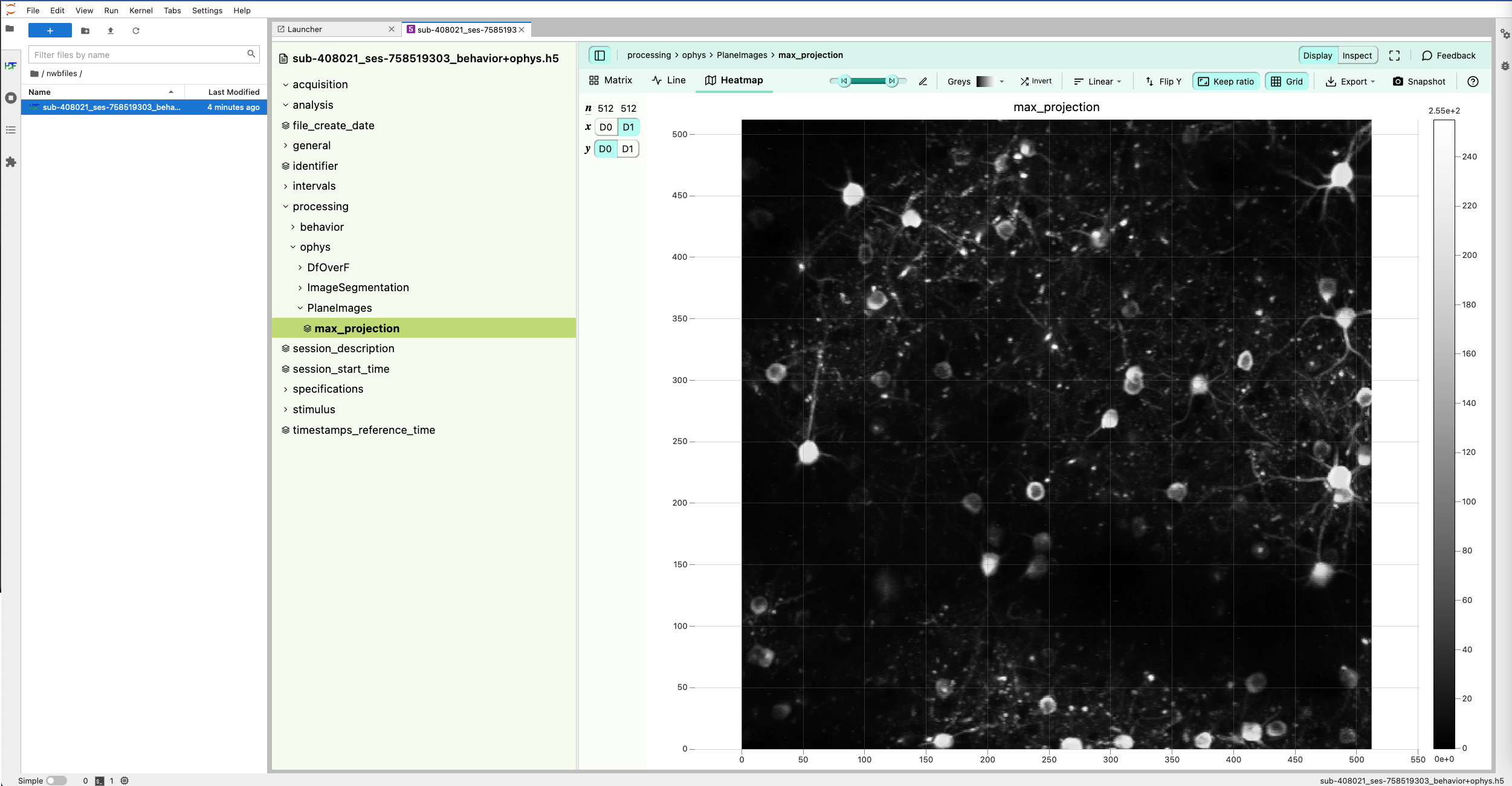
HDF Tools
A broad range of useful tools for inspecting and browsing HDF5 files, including HDFView, HDF5 plugins for Jupyter or VSCode, and command-line tools.Extracellular Electrophysiology Tools
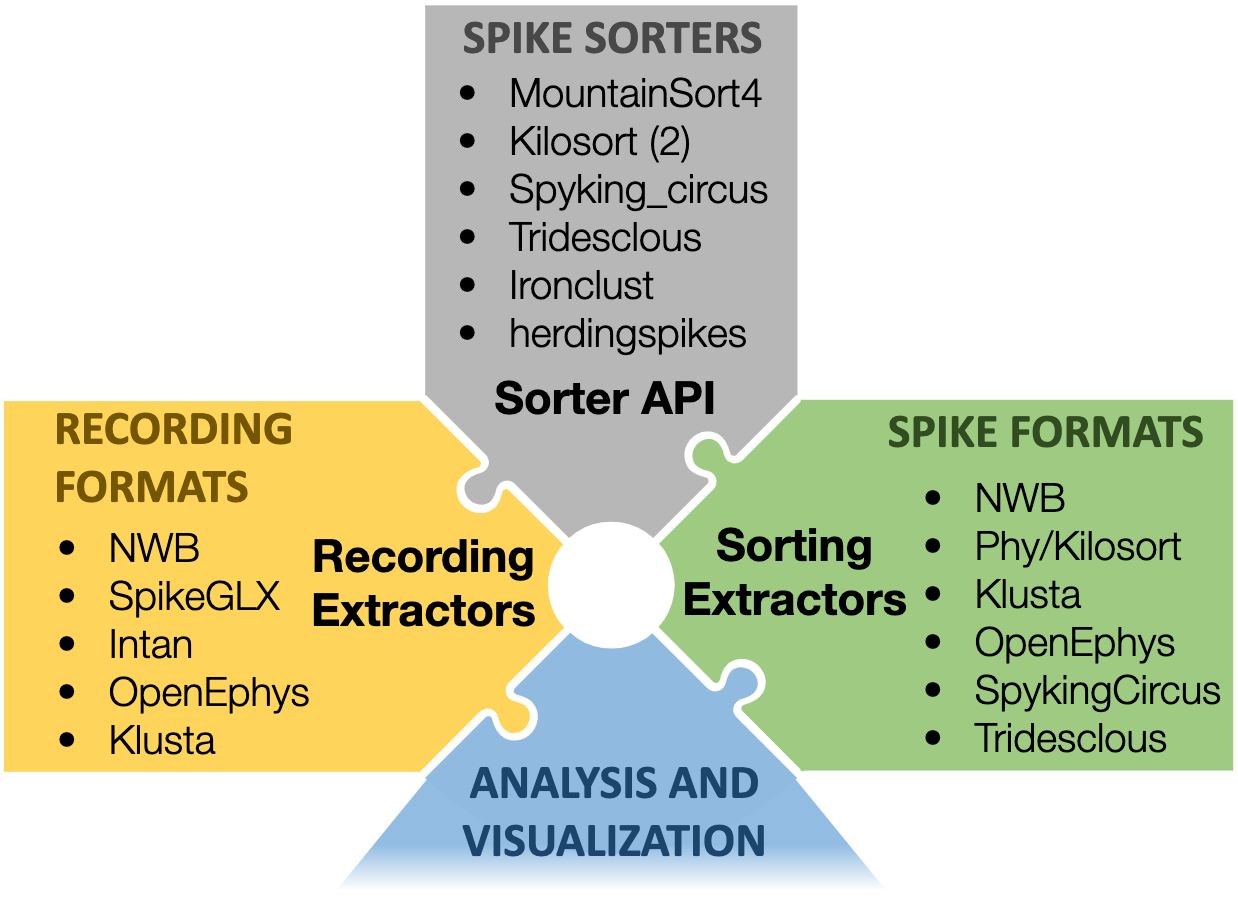
SpikeInterface
A collection of Python modules designed to improve the accessibility, reliability, and reproducibility of spike sorting and all its associated computations.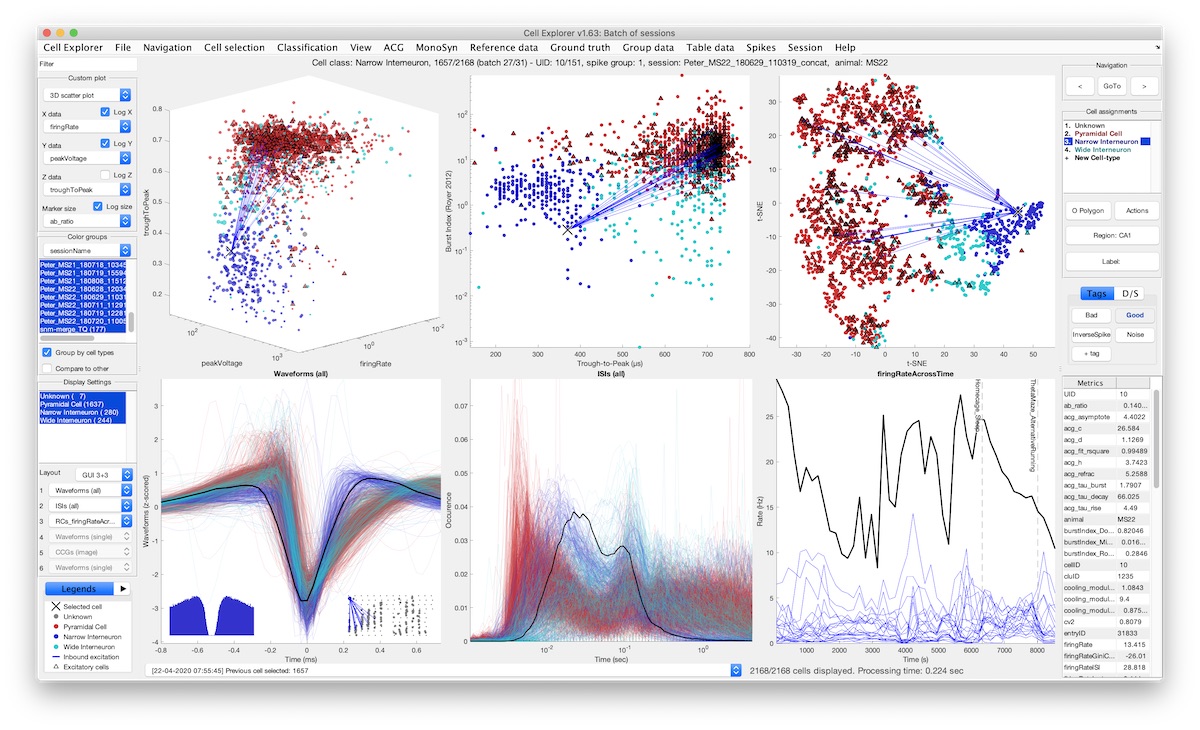
CellExplorer
A graphical user interface (GUI), a standardized processing module and data structure for exploring and classifying single cells acquired using extracellular electrodes.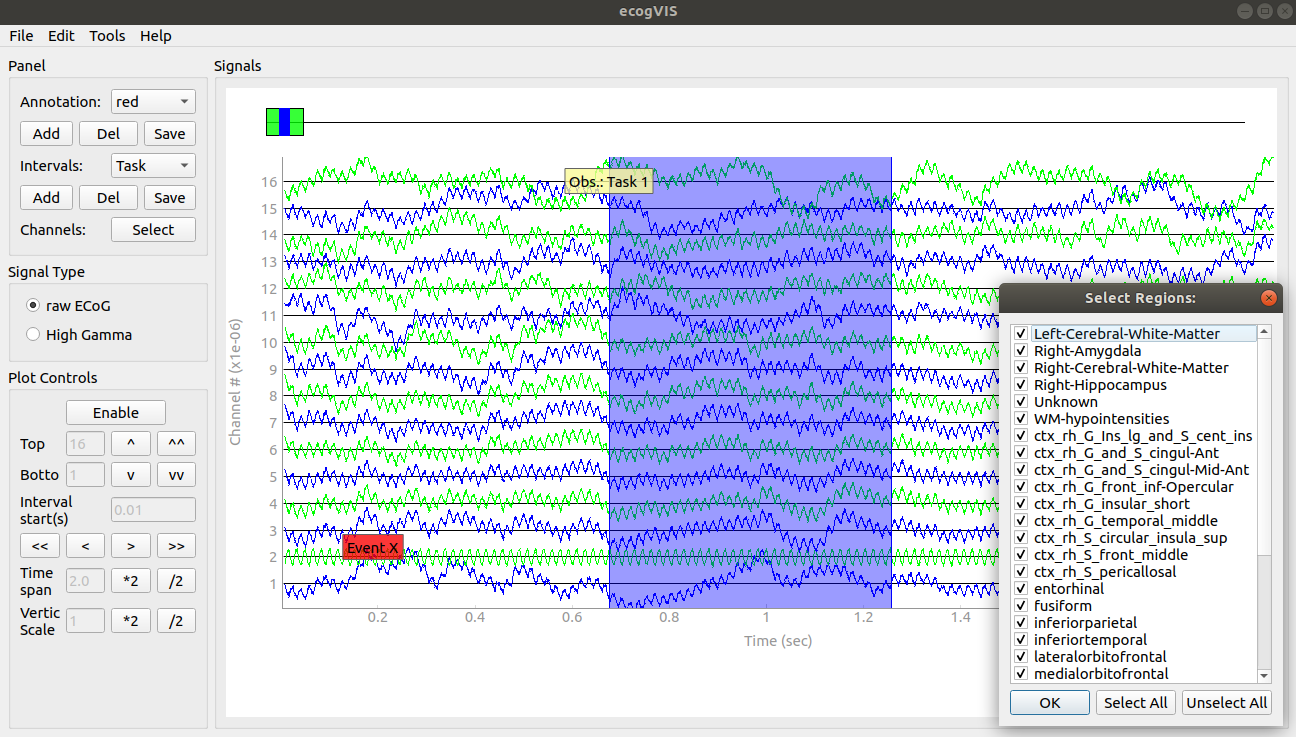
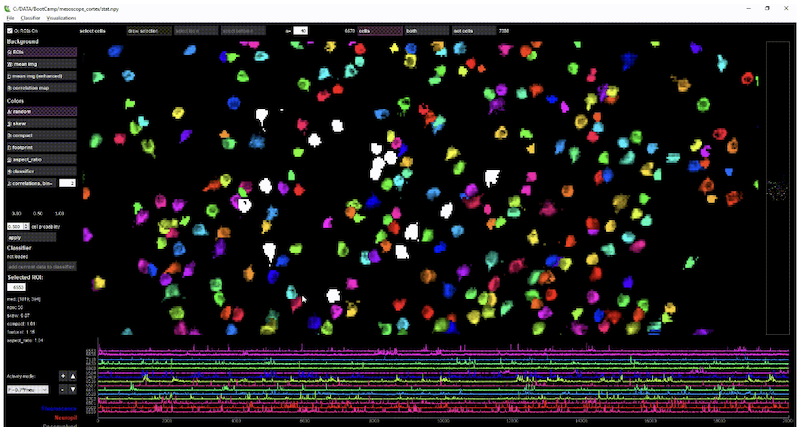
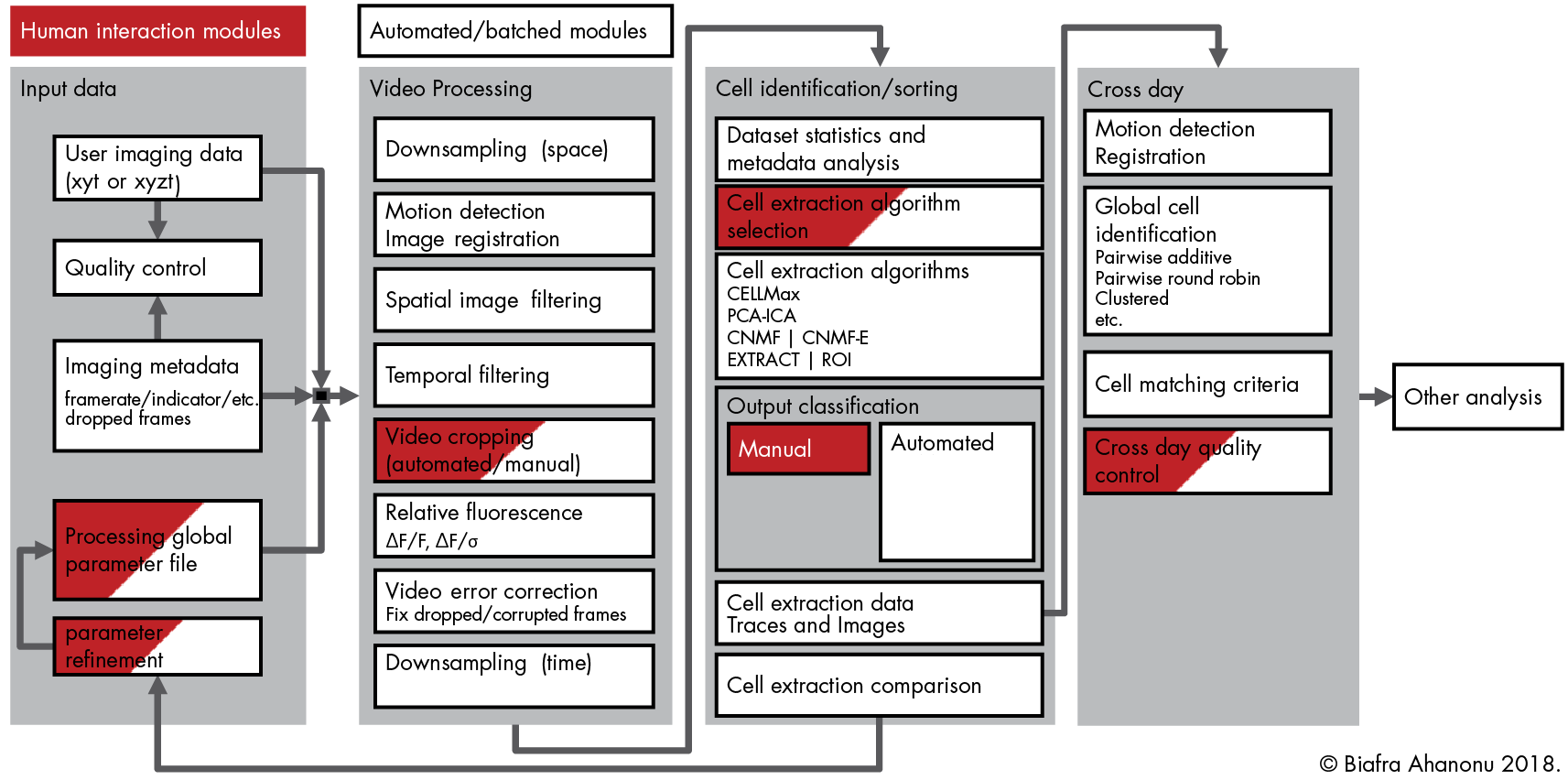
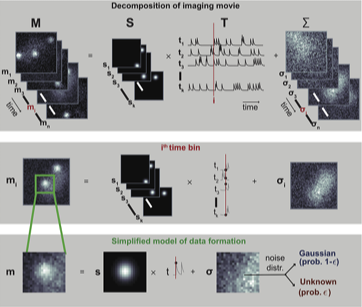
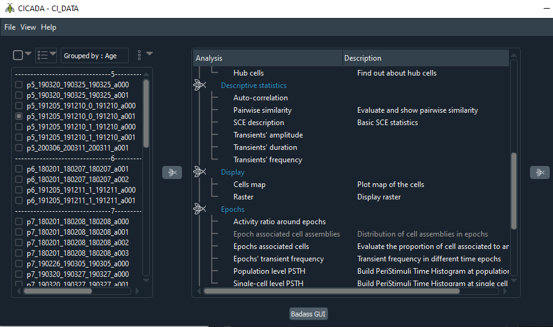
Optical Physiology Tools


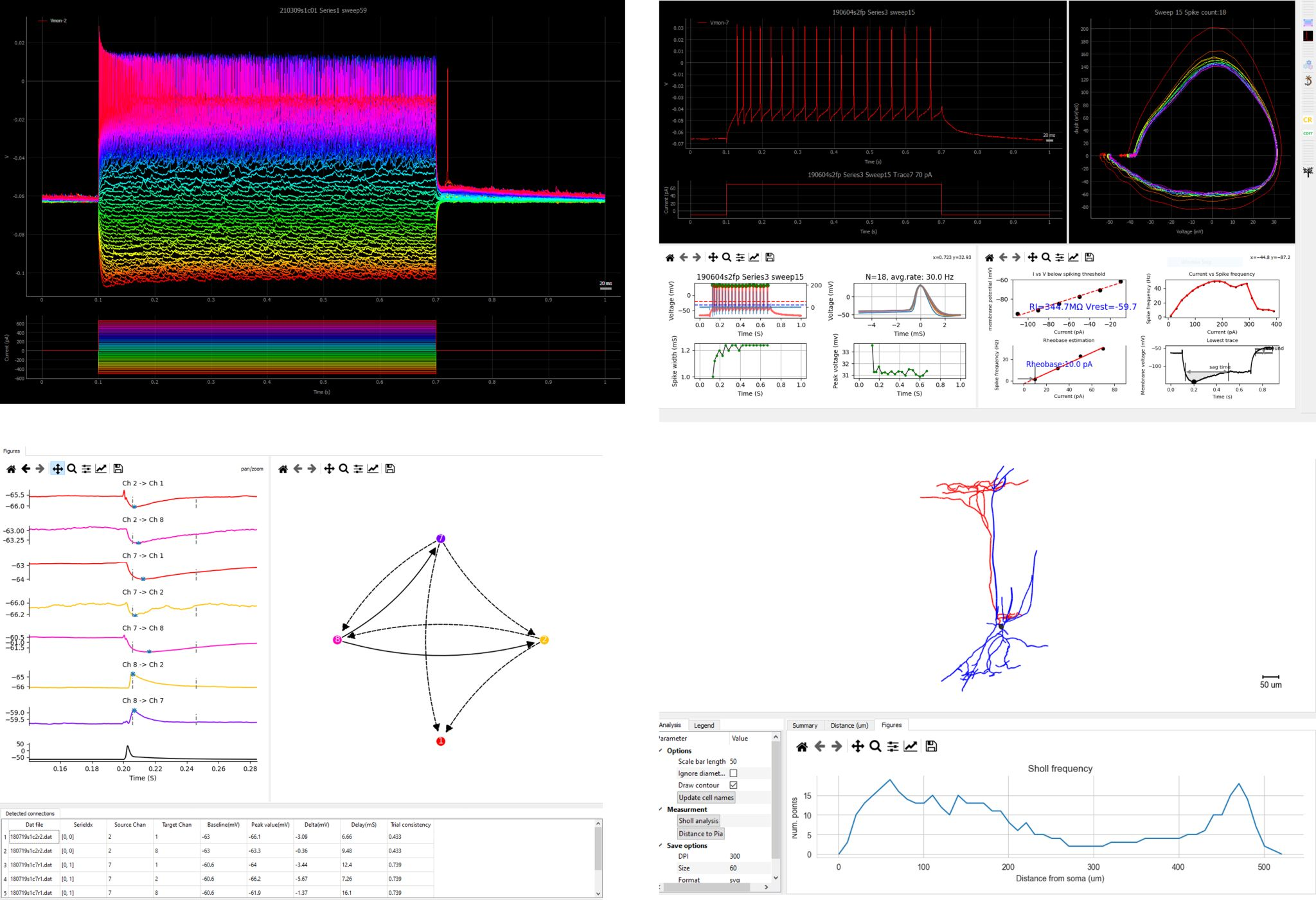
Intracellular Electrical Physiology Tools
Behavior
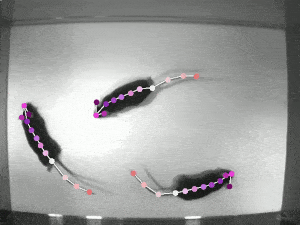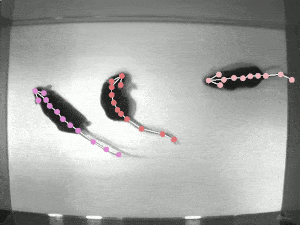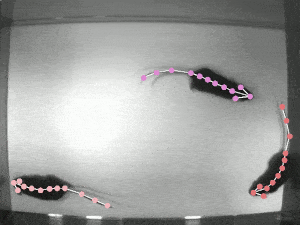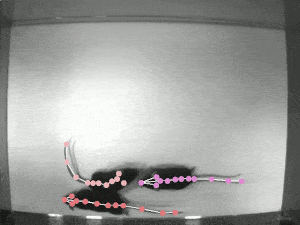
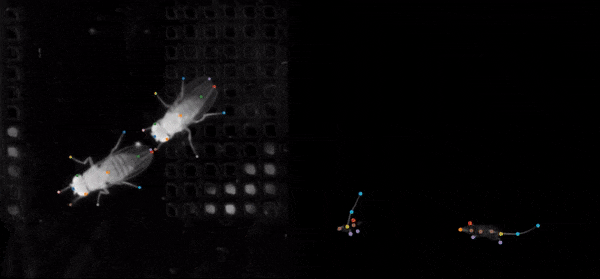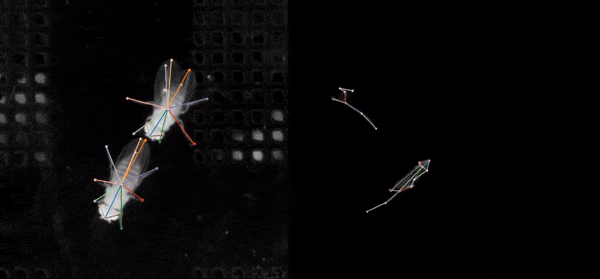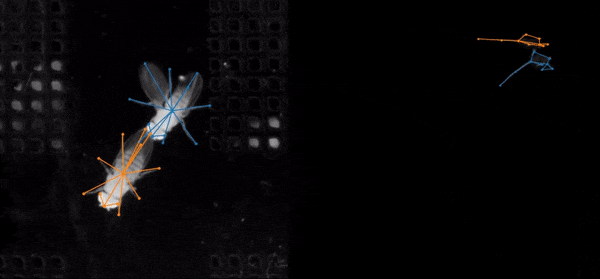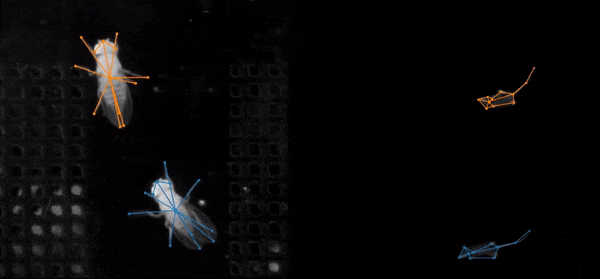
DeepLabCut
An efficient method for 2D and 3D markerless pose estimation based on transfer learning with deep neural networks that achieves excellent results with minimal training data.Data Analysis Toolbox

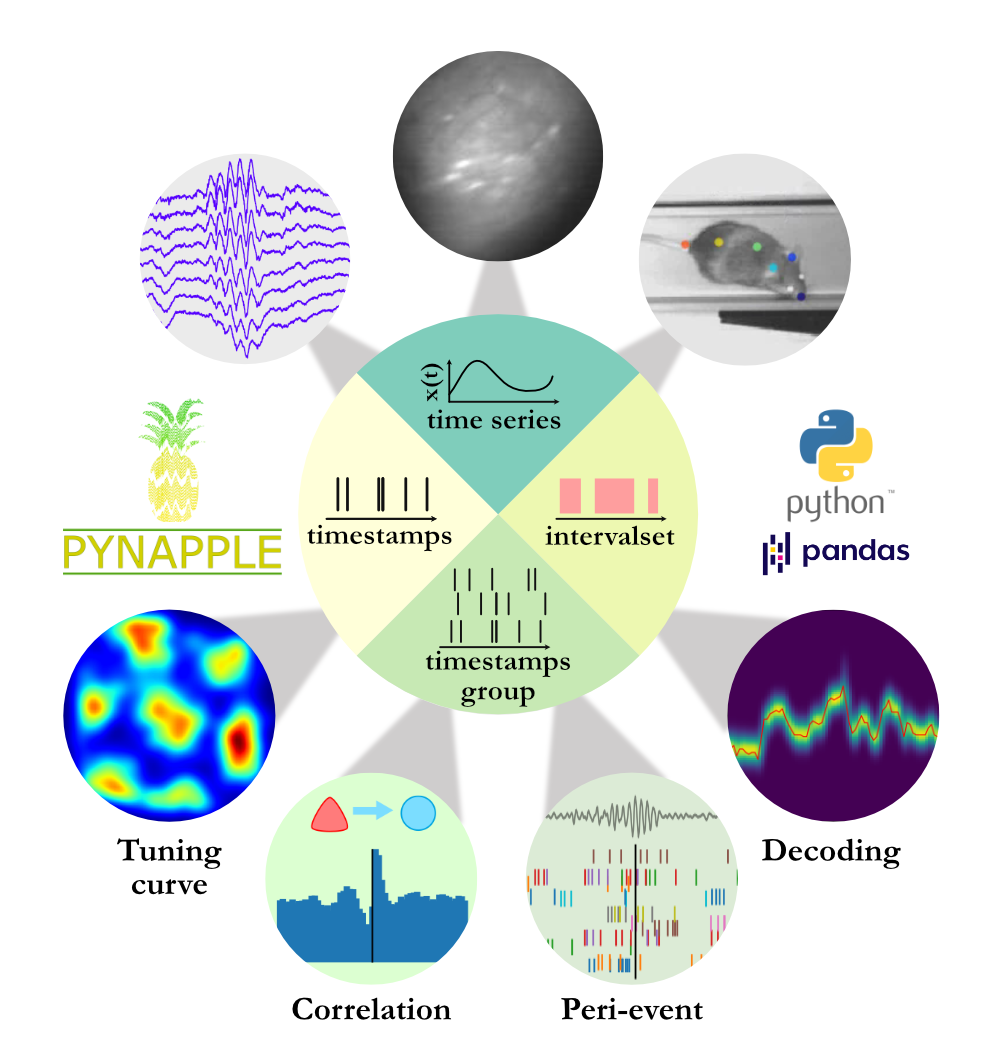
Data Archive, Publication, and Management