NWB Workshops and Hackathons
Back to Projects List
Storage of large-scale network simulation output
Key Investigators
- Kael Dai (Allen Institute) @kaeldai
- Ben Dichter (Stanford Lab) @bendichter
- Yazan Billeh (Allen Institute) @CellAssembly
Project Description
An efficent, parallizable way to store the results, and even the input, of large-scale in-silico network simulations.
Objective
- Create an extension for storing and reading discrete single unit simulation data (ie spike times from a biophysical/point network simulation).
- Create an extension for continuous data (ie membrane potential, calcium conc).
- May have to create a second extension for multi-compartment reporting (ie membrane potential along all sections of the dentrites)
Approach and Plan
Spike time recordings.
Goals: We want to be able to save spike trains from a simulation of anywhere between hundreds to millions of different cells. The previous way to do this was given every cell their own dataset.
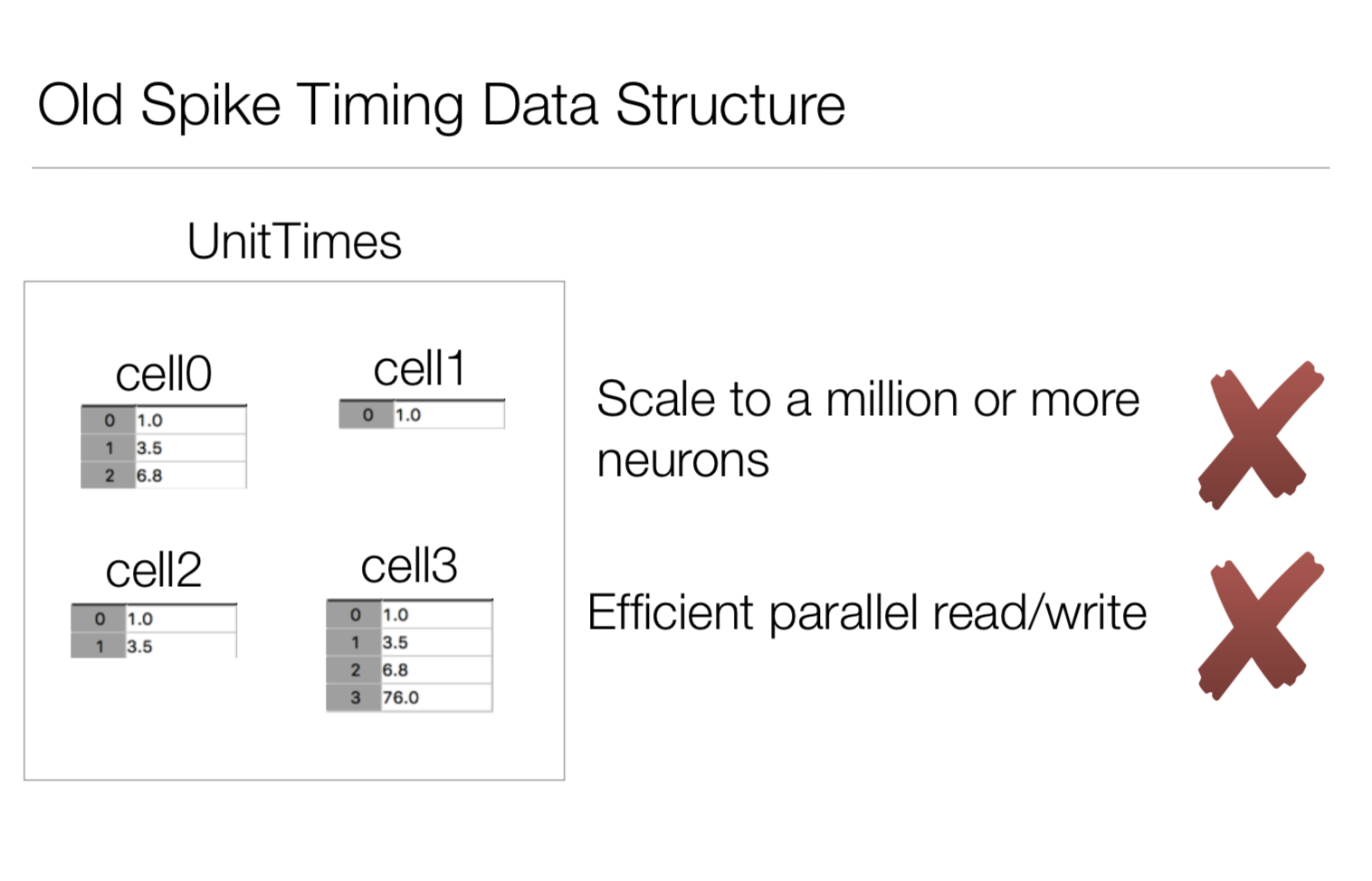
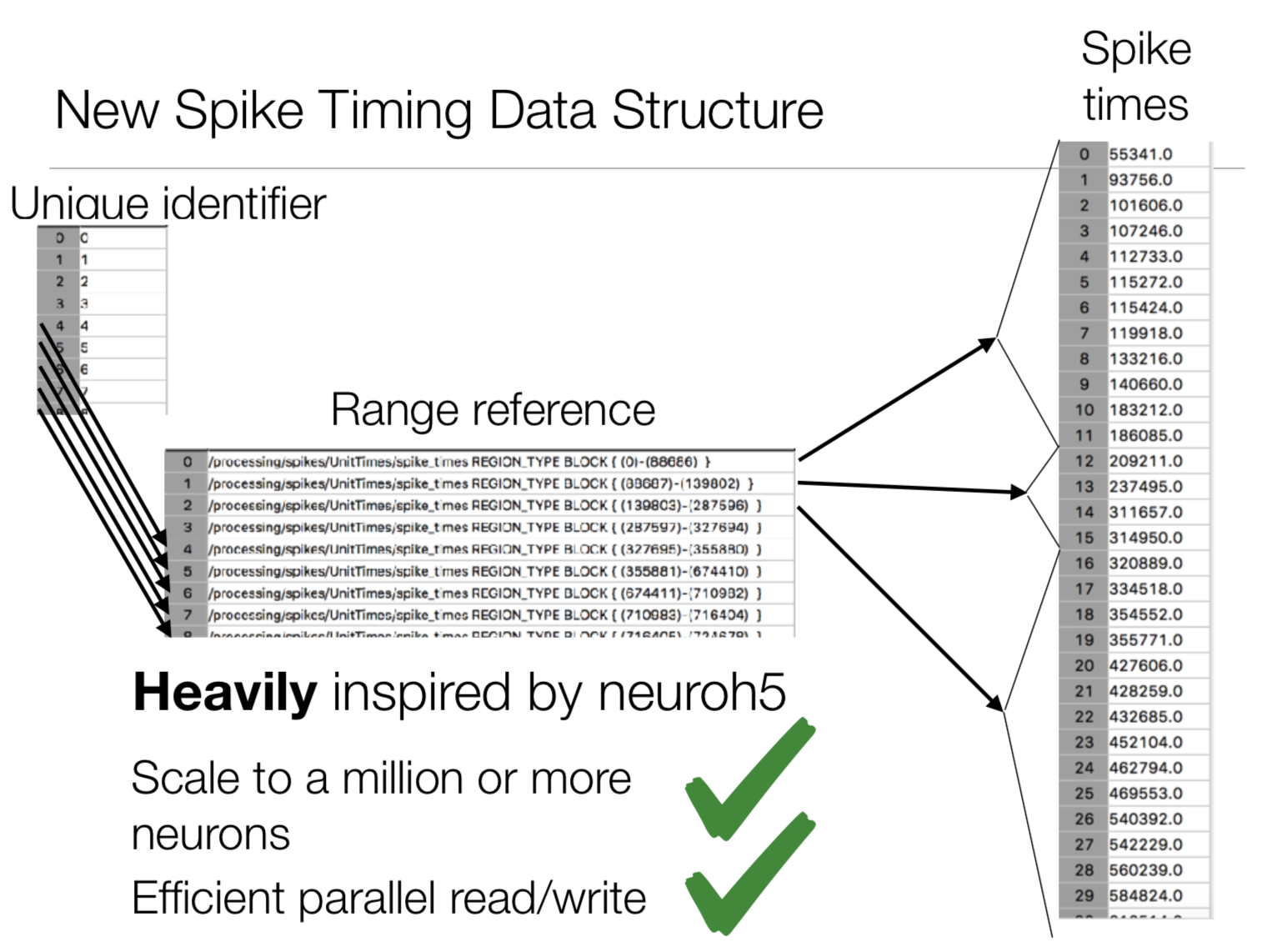
While simple to understand, having to open a dataset handle for every possible cell in the network didn’t scale well. For the new format, we store all spikes in a single file, using an index table link a given spike time with a cell gid.
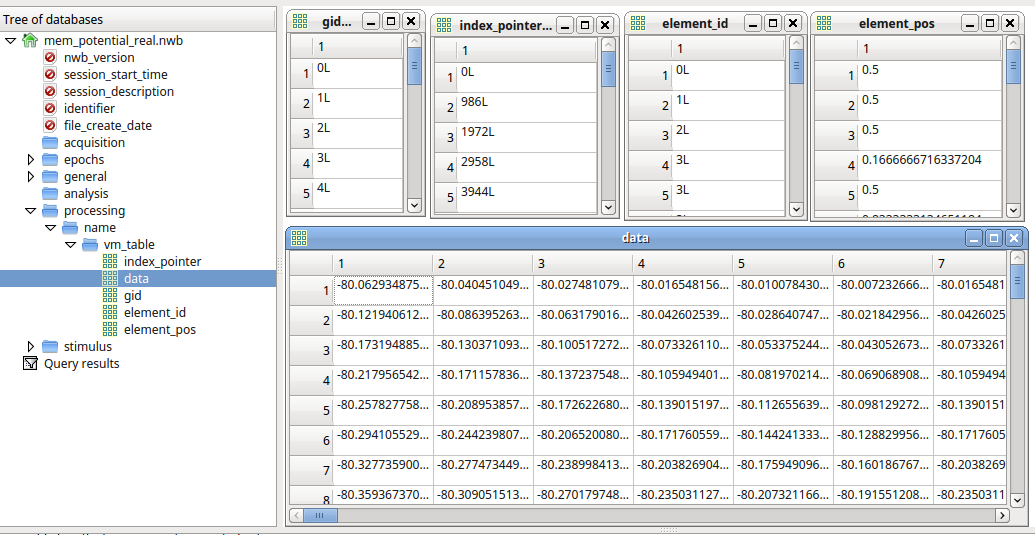
Multicell, multicompartment continuous data storage
Goals:
- Store cell variable data (membrane potential, [Ca++], etc), collected during a simulation
- Multiple cells
- Individual cells may be made up of different sections (soma, axon, dendritic branches) potentially needing to be stored.
- Want to be able to write/read in parallel.
- Want to be able to chunk data by time or by cell
Conceptural Framework:
- Index table stores range of segment ids.
- Stores relative recording position of each cell segment.
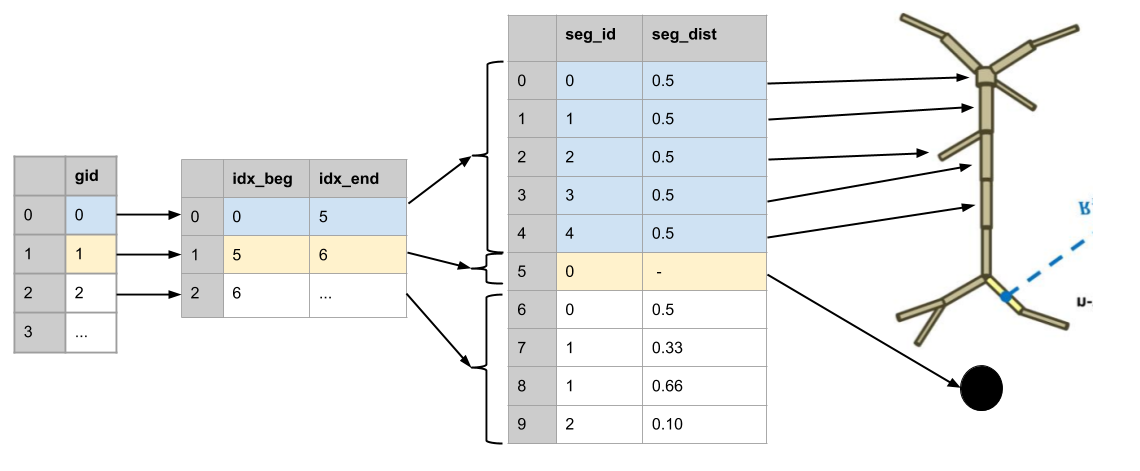
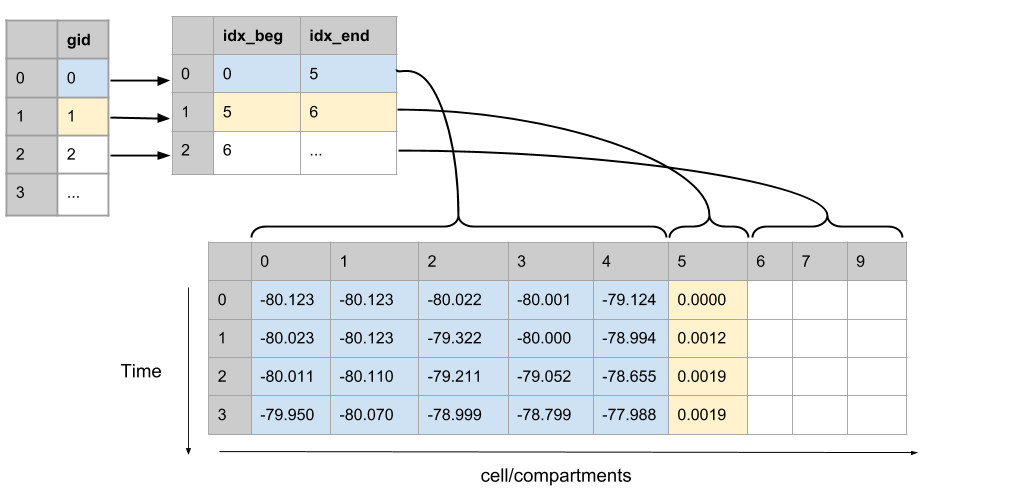
Segment recordings from each cell are stored in a single TimeSeries
Progress and Next Steps
-
Need to be able to use region references as a more explict way of indexing tables.
-
Critical: we need to be able to preallocate memory blocks and write in chunks (and to take advantage of MPI).
Illustrations
Background and References
- Forum: https://github.com/orgs/NeurodataWithoutBorders/teams/networkoutput
- Source code: https://github.com/NeurodataWithoutBorders/nwb_hackathons/tree/master/HCK04_2018_Seattle/Projects/NetworkOutput